Assignment 3¶
Problem 1¶
The goal of this exercise is to compare clustering coefficient and the length of shorthest path of Watts-Strogatz model with Erdos-Renyi model. To do so, we will create multiple instances of Watts-Strogatz model with \(N=100\) vertices and different values for \(k\). Similarly, we will generate multiple instances of ER model with \(N=100\) and \(m = \frac{k\cdot N}{2}\).
Firstly, we will import the necessary packages.
import sys, os
sys.path.insert(0, '../src/')
import algorithms.watts_strogatz as ws
import algorithms.erdos_renyi as er
import numpy as np
import matplotlib.pyplot as plt
N, beta = 100, 0.5
K = np.arange(30, 90, 2)
er_graphs = []
ws_graphs = []
for k in K:
m = k*N // 2
ws_graphs.append(er.er_nm(N, m))
er_graphs.append(ws.watts_strogatz(N, k, beta, seed=1234))
Now we will make two different plots. In one, we will compare the diameters of the above generated graphs and in the second one, we will compare clustering coefficient.
# on x axis, plot k
# on y axis, plot the diameter
ws_diam, er_diam, ws_cc, er_cc = [], [], [], []
for g1, g2 in zip(ws_graphs, er_graphs):
ws_cc.append(g1.global_clustering_coefficient())
paths = g1.shortest_path()
max_path = [[(0 if np.isinf(paths[u][v]) else paths[u][v]) for v in g1.vertices()] for u in g1.vertices()]
ws_diam.append(max(max(max_path)))
er_diam.append(g2.diameter())
er_cc.append(g2.global_clustering_coefficient())
fig = plt.figure(figsize = (15, 8))
plt.subplot(211)
er_line, = plt.plot(K, er_diam, 'b', label="Erdos Renyi diameter")
ws_line, = plt.plot(K, ws_diam, 'r', label="Watts-Strogatz diameter")
plt.legend([er_line, ws_line])
plt.xlabel("m")
plt.ylabel("diameter")
plt.subplot(212)
cc_er_line, = plt.plot(K, er_cc, 'b', label="Erdos Renyi clust coef")
cc_ws_line, = plt.plot(K, ws_cc, 'r', label="Watts-Strogatz clust coef")
plt.legend([cc_er_line, cc_ws_line])
plt.xlabel("m")
plt.ylabel("clust coef")
plt.show()
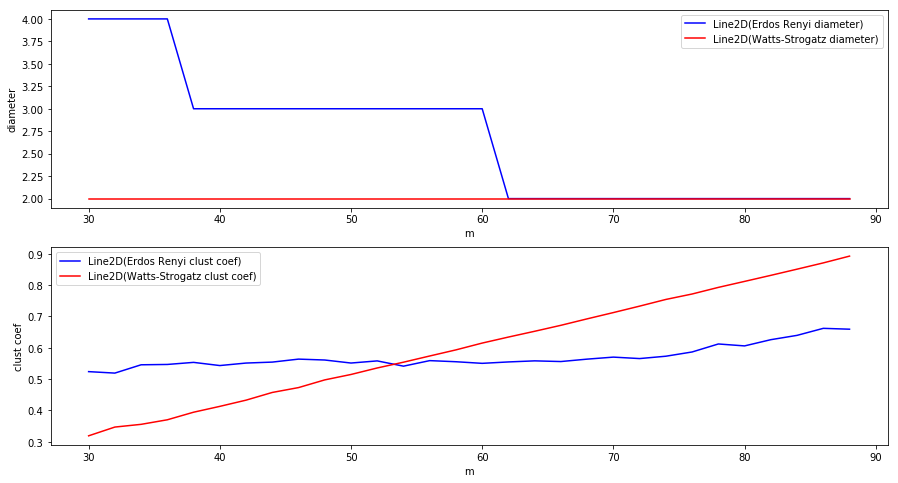
As we can see from the above plots, graphs following BA model have approximately the same diameter as ER graphs while the clustering coefficient is increased.
Problem 2¶
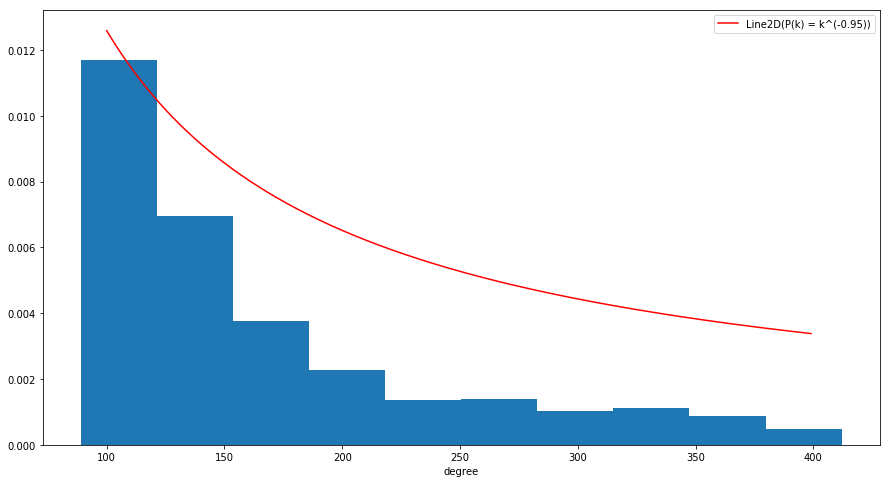
The goal of this exercise is to show that Barabasi-Albert model produces a scale free network. To do that, we will generate a random graph following BA model and compute the degree distribution. Then, we will try to approximate the obtained distribution with power law distribution.
import algorithms.barabasi_albert as ba
reload(ba)
N, m, gamma = 1000, 100, -2
M = np.arange(90, 150)
g = ba.barabasi_albert(N, m)
# ba_graphs = [ba.barabasi_albert(N, m) for m in M]
gamma = -0.95
fig = plt.figure(figsize=(15, 8))
degree_seq = np.array(g.degree_sequence(), dtype=np.float32)
degree_dist = degree_seq / sum(degree_seq)
plt.hist(degree_seq, normed=True)
degree_range = np.arange(100, 400, dtype=np.float32)
p_law = [k**gamma for k in degree_range]
pl_line, = plt.plot(degree_range, p_law, 'r-', label="P(k) = k^(-0.95)")
plt.legend([pl_line])
plt.xlabel("degree")
plt.show()